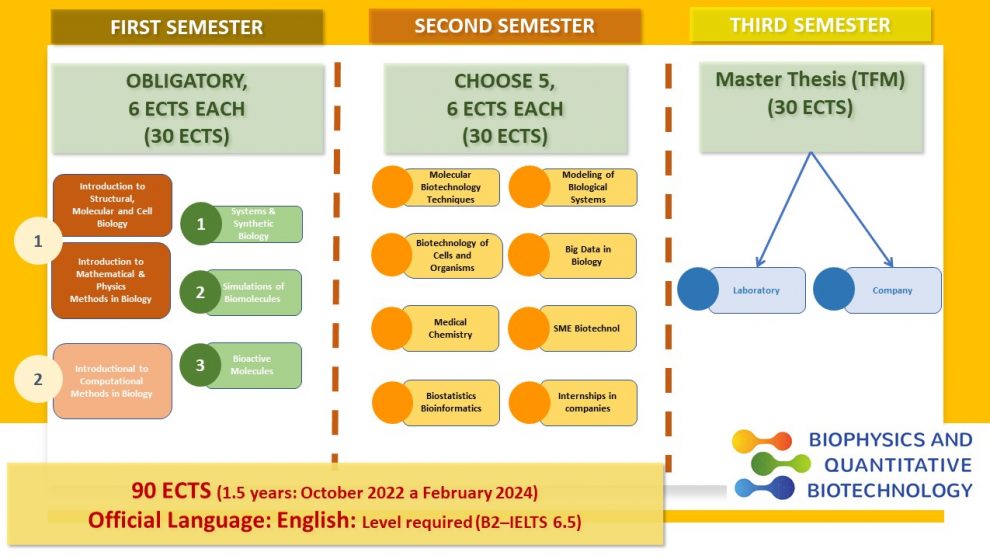
1ST SEMESTER
6 ECTS EACH (30 ECTS)
- Brief introduction to the diversity of living beings. The evolution of the species.
- Biomolecules.
- Cell Structure and physiology.
- The prokaryotic cell.The eukaryotic cell.
- Membranes and transport among compartments.
- Metabolism.
- The nucleus.
- The cell cycle: mechanisms of cell division.
- Flow of genetic information (replication, transcription and translation).
- Viruses.
- Structure databases. PDB. Bank of nucleic acid structures.
- Visualization of three-dimensional structures of macromolecules.
- The conformational stability of proteins. Interactions that contribute to protein stability. Cooperativity.
- Protein folding. Anfinsen experiment. Levinthal's paradox. Folding stages. The 'new vision' of folding. The problem of aggregation and chaperones.
- Stability and folding of nucleic acids. Stability and thermal denaturation of the double helix. Stability of folded RNA molecules. Nucleic acid folding.
- Interactions between macromolecules. Nature of the interaction surfaces. Forces involved in the interaction. Protein-protein interactions. Protein-nucleic acid interactions. Protein-lipid interactions. Protein-carbohydrate interactions. Examples.
- Biomolecules.
- Cell Structure and physiology.
- The prokaryotic cell.The eukaryotic cell.
- Membranes and transport among compartments.
- Metabolism.
- The nucleus.
- The cell cycle: mechanisms of cell division.
- Flow of genetic information (replication, transcription and translation).
- Viruses.
- Structure databases. PDB. Bank of nucleic acid structures.
- Visualization of three-dimensional structures of macromolecules.
- The conformational stability of proteins. Interactions that contribute to protein stability. Cooperativity.
- Protein folding. Anfinsen experiment. Levinthal's paradox. Folding stages. The 'new vision' of folding. The problem of aggregation and chaperones.
- Stability and folding of nucleic acids. Stability and thermal denaturation of the double helix. Stability of folded RNA molecules. Nucleic acid folding.
- Interactions between macromolecules. Nature of the interaction surfaces. Forces involved in the interaction. Protein-protein interactions. Protein-nucleic acid interactions. Protein-lipid interactions. Protein-carbohydrate interactions. Examples.
OR
Mathematical methods:
1. Introduction to linear algebra:
- Vector spaces and linear applications
- Matrices
- Eigenvalues and eigenvectors. Diagonalizability
2. Introduction to calculus
- Functions of one and several variables
- Continuity and differentiability. Taylor series.
- Riemann integral
- Systems of differential equations. Qualitative methods.
- Equations in partial derivatives.
3. Introduction to Statistics
· Basic Concepts of probability: distributions of basic probability
· Descriptive statistics.
· Basic inference: Estimate and Test of basic hypotheses.
Elements of Physics:
- Classic Mechanics of Particle Systems
- Introduction to Thermodynamics and Statistical Mechanics.
· Basic thermodynamic functions: energy and entropy.
· The concept of thermodynamic ensemble
· Microcanonical and canonical. The concept of temperature.
- Introduction to Quantum Mechanics
- The concept of wave function.
- The Schrödinger equation
- The hydrogen atom.
1. Introduction to linear algebra:
- Vector spaces and linear applications
- Matrices
- Eigenvalues and eigenvectors. Diagonalizability
2. Introduction to calculus
- Functions of one and several variables
- Continuity and differentiability. Taylor series.
- Riemann integral
- Systems of differential equations. Qualitative methods.
- Equations in partial derivatives.
3. Introduction to Statistics
· Basic Concepts of probability: distributions of basic probability
· Descriptive statistics.
· Basic inference: Estimate and Test of basic hypotheses.
Elements of Physics:
- Classic Mechanics of Particle Systems
- Introduction to Thermodynamics and Statistical Mechanics.
· Basic thermodynamic functions: energy and entropy.
· The concept of thermodynamic ensemble
· Microcanonical and canonical. The concept of temperature.
- Introduction to Quantum Mechanics
- The concept of wave function.
- The Schrödinger equation
- The hydrogen atom.
AND
Basic Computing.
- Computers, their elements and basic operation.
- Representation and coding of the information.
- Notions of Operating Systems.
- Computer networks, generalities.
Basic programming elements in some advanced language: common concepts and differences between platforms:
- Data Structures.
- Control Structures.
- Functions.
- Implementation of simple programs and algorithms.
Examples of numerical methods relevant to computational biology.
- Computers, their elements and basic operation.
- Representation and coding of the information.
- Notions of Operating Systems.
- Computer networks, generalities.
Basic programming elements in some advanced language: common concepts and differences between platforms:
- Data Structures.
- Control Structures.
- Functions.
- Implementation of simple programs and algorithms.
Examples of numerical methods relevant to computational biology.
- Introduction to systems biology: basic concepts, biological networks, data sources.
- Analysis of simple, natural and synthetic regulatory circuits. Structural description and dynamic evolution.
- Complex networks. Structural properties. Patterns.
- Regulatory and metabolic networks: structures and mechanisms of control. Dynamics.
- Study of relevant examples.
- Models of cellular systems.
- Analysis of simple, natural and synthetic regulatory circuits. Structural description and dynamic evolution.
- Complex networks. Structural properties. Patterns.
- Regulatory and metabolic networks: structures and mechanisms of control. Dynamics.
- Study of relevant examples.
- Models of cellular systems.
- Experimental determination of biomolecular structures.
- Force field descriptions for biomolecules (Molecular Mechanics, MM).
- Molecular Dynamic Simulations (MDS).
- Conformational sampling using Monte Carlo methods.
- Analysis of Molecular Dynamic Simulations (PCA, normal modes, free energies…).
- Quantum mechanics (QM) in simulations of enzymatic reactions.
- Molecular Dynamics in hybrid methods QM/MM.
- Docking methods, protein-ligand and protein-protein (or DNA).
- Biomolecular simulations in Advanced Computation environments.
- Force field descriptions for biomolecules (Molecular Mechanics, MM).
- Molecular Dynamic Simulations (MDS).
- Conformational sampling using Monte Carlo methods.
- Analysis of Molecular Dynamic Simulations (PCA, normal modes, free energies…).
- Quantum mechanics (QM) in simulations of enzymatic reactions.
- Molecular Dynamics in hybrid methods QM/MM.
- Docking methods, protein-ligand and protein-protein (or DNA).
- Biomolecular simulations in Advanced Computation environments.
- Thermodynamics of equilibrium and biomolecular interactions.
- Development of identification bioassays.
- Experimental and virtual screening of chemical libraries.
- Analysis of natural products.
- Toxicity, bioavailability and effectiveness tests.
- Structure / activity relationships.
- Tools for design and engineering of proteins,
-Stabilization, optimization and design of proteins,
- Development of identification bioassays.
- Experimental and virtual screening of chemical libraries.
- Analysis of natural products.
- Toxicity, bioavailability and effectiveness tests.
- Structure / activity relationships.
- Tools for design and engineering of proteins,
-Stabilization, optimization and design of proteins,
2ND SEMESTER
CHOOSE 5, 6 ECTS EACH (30 ECTS)
- UV-vis absorption spectroscopy: instrumentation and applications. Differential Spectroscopy. Applications in Quality Control, Biotechnology and Biomedicine.
- Spectroscopy of circular dichroism. Interpretation of CD spectra of biological samples; applications. Stability and quality control of biomolecules.
- Emission spectroscopy. Fluorescence anisotropy. FRET. Fluorescence Correlation Spectroscopy (FCS). Single particle techniques. Interpretation of spectra of biological samples. Imaging. Applications to Quality Control, Biotechnology and Biomedicine.
- Spectroscopy and rapid kinetic techniques: laser pulse-induced kinetic spectrometry and rapid mixing with stopped flow. Pre-stationary state: kinetic and interaction parameters.
- Static and Dynamic light scattering. Biotechnological applications.
- Differential scanning calorimetry (DSC). Fundamentals. Applications to protein stability and cooperativity, and to interaction affinity. DSC in formulation and quality control.
- Isothermal titration calorimetry (ITC). Fundamentals. Applications to Cooperative interaction phenomena. ITC in formulation and quality control in the biotechnology industry.
- Mass spectrometry. Mass spectrometers and their components: ionization sources and analyzers. Interpretation of mass spectra. Applications in proteomics and metabolomics.
- Optical biosensors based on Surface Plasmon Resonance (SPR). Interaction of biomolecules in real time. Design of an SPR experiment. Applications.
- Biophysical techniques in Enzymology. Enzyme activity. Inhibitors. Function activators and rescuers.
- Structural techniques for biomolecules. X-ray. NMR. Electronic cryomicroscopy.
- Instrumental equipment forspecific tasks. Miniaturized and microfluidic systems.
- Spectroscopy of circular dichroism. Interpretation of CD spectra of biological samples; applications. Stability and quality control of biomolecules.
- Emission spectroscopy. Fluorescence anisotropy. FRET. Fluorescence Correlation Spectroscopy (FCS). Single particle techniques. Interpretation of spectra of biological samples. Imaging. Applications to Quality Control, Biotechnology and Biomedicine.
- Spectroscopy and rapid kinetic techniques: laser pulse-induced kinetic spectrometry and rapid mixing with stopped flow. Pre-stationary state: kinetic and interaction parameters.
- Static and Dynamic light scattering. Biotechnological applications.
- Differential scanning calorimetry (DSC). Fundamentals. Applications to protein stability and cooperativity, and to interaction affinity. DSC in formulation and quality control.
- Isothermal titration calorimetry (ITC). Fundamentals. Applications to Cooperative interaction phenomena. ITC in formulation and quality control in the biotechnology industry.
- Mass spectrometry. Mass spectrometers and their components: ionization sources and analyzers. Interpretation of mass spectra. Applications in proteomics and metabolomics.
- Optical biosensors based on Surface Plasmon Resonance (SPR). Interaction of biomolecules in real time. Design of an SPR experiment. Applications.
- Biophysical techniques in Enzymology. Enzyme activity. Inhibitors. Function activators and rescuers.
- Structural techniques for biomolecules. X-ray. NMR. Electronic cryomicroscopy.
- Instrumental equipment forspecific tasks. Miniaturized and microfluidic systems.
- Common cell and animal models in research: bacteria, yeasts, eukaryotic cells, mice.
- Prokaryotic and eukaryotic cell culture techniques. Bacteria, yeasts. Culture of adherent cells and cells in suspension. Three-dimensional cell culture. Embryoid bodies, neurospheres, organoids. Cell culture by continuous perfusion. Microfluidic chips: design and utilities.
- Determination of metabolic parameters. Photosynthesis and cellular respiration.
- Advanced cytometry and spectrometry.
- Advanced microscopy. Multidimensional light microscopy, confocal microscopy, single particle microscopy, electron microscopy, atomic force microscopy.
- Genetic manipulation. Transformation, transfection, viral transduction. Genetic editing using CRISPR. Antisense RNA technology. Animal transgenesis and embryo manipulation. Advanced techniques for genetic manipulation in microorganisms.
- Nanoparticles in cellular biotechnology and organism. Synthesis, functionalization, administration, applications.
- Stem cells and regenerative medicine. Pluripotent embryonic and induced stem cells, targeted differentiation, cell therapy, and regenerative medicine.
- Adipocyte differentiation.
- Ultracentrifugation.
- Analysis of microscopy images.
- Functional assessment in animals. Advanced fluorescence and luminescence techniques in animals. Methods for behavioral studies in animals.
- Prokaryotic and eukaryotic cell culture techniques. Bacteria, yeasts. Culture of adherent cells and cells in suspension. Three-dimensional cell culture. Embryoid bodies, neurospheres, organoids. Cell culture by continuous perfusion. Microfluidic chips: design and utilities.
- Determination of metabolic parameters. Photosynthesis and cellular respiration.
- Advanced cytometry and spectrometry.
- Advanced microscopy. Multidimensional light microscopy, confocal microscopy, single particle microscopy, electron microscopy, atomic force microscopy.
- Genetic manipulation. Transformation, transfection, viral transduction. Genetic editing using CRISPR. Antisense RNA technology. Animal transgenesis and embryo manipulation. Advanced techniques for genetic manipulation in microorganisms.
- Nanoparticles in cellular biotechnology and organism. Synthesis, functionalization, administration, applications.
- Stem cells and regenerative medicine. Pluripotent embryonic and induced stem cells, targeted differentiation, cell therapy, and regenerative medicine.
- Adipocyte differentiation.
- Ultracentrifugation.
- Analysis of microscopy images.
- Functional assessment in animals. Advanced fluorescence and luminescence techniques in animals. Methods for behavioral studies in animals.
Drug targets and drugs
- Enzymes and receptors as pharmacological targets.
- Nucleic acids and other pharmacological targets.
- Antimicrobial agents.
- Anticancer agents.
- Drugs for the central nervous system.
- Other drugs.
Basic tools in Medical Chemistry
- Find header compounds.
- Combinatorial synthesis.
- Chiral drugs and their synthesis.
- (Q) SARs to guide Medical Chemistry.
- Computers in Medical Chemistry.
Drug characterization: pharmacokinetics and pharmacodynamics
- Characterization of the header compounds
- Pharmacokinetics and pharmacodynamics
- Safety and Toxicology. Animal models of disease.
Chemical strategies to improve drugs
- Improvement of solubility and affinity for the target.
- Improvement of chemical and metabolic stability.
- Improvement of protein binding and permeability.
- Decreased toxicity.
- Improved administration (prodrugs, encapsulation).
From the drug to the patient
- Intellectual property and patents in drug discovery
- Fundamentals of clinical trials
- Bring the drug to the market
- Illustrative examples in drug discovery
- Enzymes and receptors as pharmacological targets.
- Nucleic acids and other pharmacological targets.
- Antimicrobial agents.
- Anticancer agents.
- Drugs for the central nervous system.
- Other drugs.
Basic tools in Medical Chemistry
- Find header compounds.
- Combinatorial synthesis.
- Chiral drugs and their synthesis.
- (Q) SARs to guide Medical Chemistry.
- Computers in Medical Chemistry.
Drug characterization: pharmacokinetics and pharmacodynamics
- Characterization of the header compounds
- Pharmacokinetics and pharmacodynamics
- Safety and Toxicology. Animal models of disease.
Chemical strategies to improve drugs
- Improvement of solubility and affinity for the target.
- Improvement of chemical and metabolic stability.
- Improvement of protein binding and permeability.
- Decreased toxicity.
- Improved administration (prodrugs, encapsulation).
From the drug to the patient
- Intellectual property and patents in drug discovery
- Fundamentals of clinical trials
- Bring the drug to the market
- Illustrative examples in drug discovery
- Inference and classical statistical modeling. The Linear Model and the Generalized Linear Model.
- Bayesian modeling and inference.
- Multivariate data. Dimension reduction techniques and classification techniques
- Supervised / Unsupervised Statistical Learning Techniques. Classification and prediction models. ROC curves. Validation.
- Markov chains. Hidden Markov Models.
- Hypothesis Testing and types of errors and P-value. Multiple tests and control of error rates: Bonferroni correction, False Discovery Rate (FDR) and E-value.
- Programming oriented to data processing in bioinformatics: reading, manipulation and writing of files in the terminal and in scripts.
- Common formats in bioinformatics: nucleic acid and protein sequences and their alignments (FASTA), molecular structures (PDB) and phylogenetic trees (Newick).
- Algorithms of the dynamic programming for local and global alignments.
- Search for similar sequences in local databases by means of alignments.
- Multiple alignments of DNA and protein sequences.
- Alignments of protein structures and calculation of RMSD.
- Design, development and fundamentals of analysis of RNAseq experiments.
- Bayesian modeling and inference.
- Multivariate data. Dimension reduction techniques and classification techniques
- Supervised / Unsupervised Statistical Learning Techniques. Classification and prediction models. ROC curves. Validation.
- Markov chains. Hidden Markov Models.
- Hypothesis Testing and types of errors and P-value. Multiple tests and control of error rates: Bonferroni correction, False Discovery Rate (FDR) and E-value.
- Programming oriented to data processing in bioinformatics: reading, manipulation and writing of files in the terminal and in scripts.
- Common formats in bioinformatics: nucleic acid and protein sequences and their alignments (FASTA), molecular structures (PDB) and phylogenetic trees (Newick).
- Algorithms of the dynamic programming for local and global alignments.
- Search for similar sequences in local databases by means of alignments.
- Multiple alignments of DNA and protein sequences.
- Alignments of protein structures and calculation of RMSD.
- Design, development and fundamentals of analysis of RNAseq experiments.
-Statistical mechanics, statistical ensembles, interactions, cooperativity, phase transitions
- Biopolymer models.
- Coarse-grained models.
- Dynamical models in biologically relevant networks.
- Stochastic models: Brownian motion and diffusion.
- Langevin and Fokker-Planck equations.
- The Master Equation in Chemistry. Gillespie's Algorithm.
- Kramers theory in chemical kinetics.
- Simulation techniques: Monte-Carlo methods and applications.
- Biopolymer models.
- Coarse-grained models.
- Dynamical models in biologically relevant networks.
- Stochastic models: Brownian motion and diffusion.
- Langevin and Fokker-Planck equations.
- The Master Equation in Chemistry. Gillespie's Algorithm.
- Kramers theory in chemical kinetics.
- Simulation techniques: Monte-Carlo methods and applications.
-Main applications of data science in Bio-medicine; main types of data sets.
-Computer programming oriented to data management and visualization.
-Quality control in biomedical big-data. Identification and neutralization of batch effects and dimensionality reduction techniques.
-Multiple testing in big-data. Different approaches, and use of empirical null models.
-Functional enrichment analysis.
-Analysis of genomic sequencing datasets.
-Single-cell technologies: basic definitions, applications and data analysis.
-Applications of networks theory to -omics data.
-Basic concepts of causal inference.
-The problem of reproducibility in Biomedical data science: challenges, precautions and practices to avoid.
-Computer programming oriented to data management and visualization.
-Quality control in biomedical big-data. Identification and neutralization of batch effects and dimensionality reduction techniques.
-Multiple testing in big-data. Different approaches, and use of empirical null models.
-Functional enrichment analysis.
-Analysis of genomic sequencing datasets.
-Single-cell technologies: basic definitions, applications and data analysis.
-Applications of networks theory to -omics data.
-Basic concepts of causal inference.
-The problem of reproducibility in Biomedical data science: challenges, precautions and practices to avoid.
There will be an exhaustive sightsee of the fundamental aspects in the elaboration of a business plan and that are fundamentally related to strategic, commercial, technical and economic-financial aspects of the company. The contents are structured according to the following points:
- The concept of entrepreneur, company and entrepreneurial process.
- Innovation and its types.
- Mechanisms of appropriation of the value of innovation: patents and secrecy.
- The role of SMEs in economic activity.
- Design thinking.
- CANVAS: representation of the business model.
- The business plan and the lean start-up.
- Entrepreneurship.
- Strategic analysis of the biotech company.
- Sources of financing for SMEs.
- Legal aspects of the company: legal form, creation and taxation.
- The concept of entrepreneur, company and entrepreneurial process.
- Innovation and its types.
- Mechanisms of appropriation of the value of innovation: patents and secrecy.
- The role of SMEs in economic activity.
- Design thinking.
- CANVAS: representation of the business model.
- The business plan and the lean start-up.
- Entrepreneurship.
- Strategic analysis of the biotech company.
- Sources of financing for SMEs.
- Legal aspects of the company: legal form, creation and taxation.
- The specific contents will depend on the host company/center where the training will take place.
- The student will be assigned two tutors (one at the University and the other at the company, entity or institution) with whom he/she must meet periodically. These tutors are in charge of supervising the work carried out and assessing the learning.
-The student will prepare a notebook of the tasks and activities in which he/she participates.
- The student will be assigned two tutors (one at the University and the other at the company, entity or institution) with whom he/she must meet periodically. These tutors are in charge of supervising the work carried out and assessing the learning.
-The student will prepare a notebook of the tasks and activities in which he/she participates.
3RD SEMESTER
30 ECTS
The Master Project entails a high level of specialization on a research subject, strictly related to the master scopes, that the student chooses, according to her/his curriculum and interests, from among a wide range of topics, proposed by the faculty (mode A) and companies (mode B). The project will allow the student to come in contact to hot research topics, and master some of the experimental and computational techniques and equipments available in modern biophysics and biotechnology labs.
In mode A (oriented towards fundamental research), students will choose a project proposed and supervised by a member of the faculty of the Master programme, and related to active research lines, developed in their labs at the School of Science, at the Institute of Biocomputation and Physics of Complex System (see labs here: https://www.bifi.es/scientific-equipment/ ), or at collaborating research centers.
In mode B, oriented towards the biotechnology company, the project will be jointly supervised by a member of the faculty of the master, and by a director within the company (ideally a PhD from their RD department), on a topic proposed by the latter, according to the company interests and availability.
In mode A (oriented towards fundamental research), students will choose a project proposed and supervised by a member of the faculty of the Master programme, and related to active research lines, developed in their labs at the School of Science, at the Institute of Biocomputation and Physics of Complex System (see labs here: https://www.bifi.es/scientific-equipment/ ), or at collaborating research centers.
In mode B, oriented towards the biotechnology company, the project will be jointly supervised by a member of the faculty of the master, and by a director within the company (ideally a PhD from their RD department), on a topic proposed by the latter, according to the company interests and availability.
- Study of the behavior of repeat proteins with simple native-centric models.
- Genomic differentiation and detection of signatures of selection between strains of Iberian pigs.
- A Multiplex network approach to study the responses of M. tuberculosis to in-vitro induced stress and its relation with epitope conservation.
- Magnetic nanoparticle-based hyperthermia for cancer treatment in different biological systems.
- Molecular characterization of mutations in the Apoptosis Inducing Factor (AIF) related with neurological disorders.
- Towards the molecular characterization of the human AIFM3 (apoptosis-inducing factor, mitochondrion-associated 3).
- Analysis of a mutated SOxR protein conferring hyper-resistance to Escherichia coli.
- Quantum Optimal Control in Biophysics: studying optimization methods.
- Functional analysis of responses to stress in distant prokaryotes: comparison between Mycobacterium tuberculosis and Escherichia coli.
- Structural and functional characterization of ferredoxin-NADP+/H oxidoreductase (FPR) and FAD synthetase from Brucella ovis.
- Evaluation of the mutome associated to Rett syndrome.
- Refinement of a statistical model for antibody humanization.
- Magnetic nanoparticles immobilization on cell membranes mediated by cadherins for magnetic hyperthermia studies.
- Mode of action elucidation studies of new antimicrobial compounds.
- Conformational analysis of thioredoxin reductases by atomic force microscopy.
- Quantitative Structure Activity Studies (QSAR) in compound series with anti-microbial activities.
- Study to the changes to human Apoptosis inducing Factor (hAIF) interactions with its physiological ligands induced by mutations in its codifying gene.
- Study of the structure-propensities of the alpha-synuclein sequence.
- Current treatments for MECP2 Duplication Syndrome.
- In silico modelling of single-cell fibroblast migration in response to 3D chemical gradients.
- Computational modeling of the folding energy of proteins with three experimentally characterized states.
- Characterization of molecular heterogeneity in celiac disease patients.
- Towards the calculation of protein stability in wild type and mutant proteins associated to Rett syndrome.
- Development and validation of real-time PCR assays for the quantification of pathogenic human viruses with diagnostic purposes.
- Testing a model for the calculation of protein stability: Application on mutations associated to genetic disorders.
- Application of a statistical inference model to the prediction of antibody affinity from sequence analysis.
- Modeling inter-individual variation in transcriptional coherence in single-cell RNA-seq data 3D co-culture of tumor spheroids and fibroblasts in microfluidic devices.
- Co-cultured neurons and microglia treated with neuromelanin as a model to study the events leading to protein aggregation in Parkinson ́s disease.
- Development of a HTS assay for inhibitors against GalNAc-T11.
- Chanelling of FMN from Homo sapiens Riboflavin kinase to Pyridoxine 5'-phosphate oxidase.
- Study of the translocation and degradation of repeat proteins by the protein complex clpXP.
- Characterisation of the possible interaction between alpha -synucleinanda tau and the formation of biomolecular condensates of both proteins which could lead to the formation of amyloid aggregates.
- BFT3 virtual screening: combining experimental and computational tools.
- SARS-COV-2 3CLpro virtual screening: combining experimental and computational tools.
- Genomic differentiation and detection of signatures of selection between strains of Iberian pigs.
- A Multiplex network approach to study the responses of M. tuberculosis to in-vitro induced stress and its relation with epitope conservation.
- Magnetic nanoparticle-based hyperthermia for cancer treatment in different biological systems.
- Molecular characterization of mutations in the Apoptosis Inducing Factor (AIF) related with neurological disorders.
- Towards the molecular characterization of the human AIFM3 (apoptosis-inducing factor, mitochondrion-associated 3).
- Analysis of a mutated SOxR protein conferring hyper-resistance to Escherichia coli.
- Quantum Optimal Control in Biophysics: studying optimization methods.
- Functional analysis of responses to stress in distant prokaryotes: comparison between Mycobacterium tuberculosis and Escherichia coli.
- Structural and functional characterization of ferredoxin-NADP+/H oxidoreductase (FPR) and FAD synthetase from Brucella ovis.
- Evaluation of the mutome associated to Rett syndrome.
- Refinement of a statistical model for antibody humanization.
- Magnetic nanoparticles immobilization on cell membranes mediated by cadherins for magnetic hyperthermia studies.
- Mode of action elucidation studies of new antimicrobial compounds.
- Conformational analysis of thioredoxin reductases by atomic force microscopy.
- Quantitative Structure Activity Studies (QSAR) in compound series with anti-microbial activities.
- Study to the changes to human Apoptosis inducing Factor (hAIF) interactions with its physiological ligands induced by mutations in its codifying gene.
- Study of the structure-propensities of the alpha-synuclein sequence.
- Current treatments for MECP2 Duplication Syndrome.
- In silico modelling of single-cell fibroblast migration in response to 3D chemical gradients.
- Computational modeling of the folding energy of proteins with three experimentally characterized states.
- Characterization of molecular heterogeneity in celiac disease patients.
- Towards the calculation of protein stability in wild type and mutant proteins associated to Rett syndrome.
- Development and validation of real-time PCR assays for the quantification of pathogenic human viruses with diagnostic purposes.
- Testing a model for the calculation of protein stability: Application on mutations associated to genetic disorders.
- Application of a statistical inference model to the prediction of antibody affinity from sequence analysis.
- Modeling inter-individual variation in transcriptional coherence in single-cell RNA-seq data 3D co-culture of tumor spheroids and fibroblasts in microfluidic devices.
- Co-cultured neurons and microglia treated with neuromelanin as a model to study the events leading to protein aggregation in Parkinson ́s disease.
- Development of a HTS assay for inhibitors against GalNAc-T11.
- Chanelling of FMN from Homo sapiens Riboflavin kinase to Pyridoxine 5'-phosphate oxidase.
- Study of the translocation and degradation of repeat proteins by the protein complex clpXP.
- Characterisation of the possible interaction between alpha -synucleinanda tau and the formation of biomolecular condensates of both proteins which could lead to the formation of amyloid aggregates.
- BFT3 virtual screening: combining experimental and computational tools.
- SARS-COV-2 3CLpro virtual screening: combining experimental and computational tools.
